Test - MT Bees
Really just testing out a workflow.
This is an .ipynb file I converted to md with jupyter nbconvert but there was still some special handling involved to get this into github pages correctly. Weird workflow.
Converting with jupyter nbconvert -to markdown [filename] produced the .md and .png files successfully but I couldn’t just mv them to the _posts dir of my stevenranney.github.io jekyll site. There was some additional handling to get the images to appear correctly. Could probably script the special handling–and I may–but for now this is okay.
My guess is that someone has probably already handled the scripting. I may dig around and find out.
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import scipy.stats as stats
from datetime import datetime, timedelta
from scipy.optimize import curve_fit
%matplotlib inline
# Seaborn defaults
sns.set_theme()
sns.set(style = 'white', font_scale = 1.5, rc = {'figure.figsize':(16, 9)})
bee = pd.read_csv('../../Google Drive/job_tech_learning/data/Bumblebeesandfl/MT_2023_Bee_Plant_Data.csv')
bee.head()
| Site ID | Date | Bee Survey Length (min) | Number of Bumble Bees Captured | Number of Plant Species in Flower | List of Plant Species in Flower | |
|---|---|---|---|---|---|---|
| 0 | x01a | 5/6/2023 | 45 | 0 | 17 | Alyssum desertorum (desert madwort), Arabis sp... |
| 1 | x01a | 5/18/2023 | 90 | 2 | 22 | Alyssum desertorum (desert madwort), Astragalu... |
| 2 | x01a | 6/28/2023 | 90 | 1 | 26 | Achillea millefolium (common yarrow), Alyssum ... |
| 3 | x01b | 4/26/2023 | 45 | 0 | 6 | Alyssum desertorum (desert madwort), Lomatium ... |
| 4 | x01b | 5/18/2023 | 90 | 0 | 28 | Allium textile (textile onion), Alyssum desert... |
bee.shape
(65, 6)
rename_cols = {
"Site ID": 'id',
"Date": 'date_str',
"Bee Survey Length (min)": "survey_length",
"Number of Bumble Bees Captured": 'n_captured',
"Number of Plant Species in Flower": 'n_flowering_plants',
"List of Plant Species in Flower": 'sp_str'
}
bee = (
bee.
rename(columns = rename_cols).
assign(
date = lambda x: pd.to_datetime(x.date_str, format = "%m/%d/%Y")
)
)
bee
| id | date_str | survey_length | n_captured | n_flowering_plants | sp_str | date | |
|---|---|---|---|---|---|---|---|
| 0 | x01a | 5/6/2023 | 45 | 0 | 17 | Alyssum desertorum (desert madwort), Arabis sp... | 2023-05-06 |
| 1 | x01a | 5/18/2023 | 90 | 2 | 22 | Alyssum desertorum (desert madwort), Astragalu... | 2023-05-18 |
| 2 | x01a | 6/28/2023 | 90 | 1 | 26 | Achillea millefolium (common yarrow), Alyssum ... | 2023-06-28 |
| 3 | x01b | 4/26/2023 | 45 | 0 | 6 | Alyssum desertorum (desert madwort), Lomatium ... | 2023-04-26 |
| 4 | x01b | 5/18/2023 | 90 | 0 | 28 | Allium textile (textile onion), Alyssum desert... | 2023-05-18 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 60 | x10c | 6/20/2023 | 90 | 0 | 19 | Achillea millefolium (common yarrow), Alyssum ... | 2023-06-20 |
| 61 | x11a | 4/23/2023 | 45 | 0 | 2 | Lomatium foeniculaceum (desert biscuitroot), P... | 2023-04-23 |
| 62 | x11b | 5/20/2023 | 90 | 0 | 11 | Allium textile (textile onion), Alyssum desert... | 2023-05-20 |
| 63 | x11b | 6/7/2023 | 90 | 2 | 17 | Achillea millefolium (common yarrow), Alyssum ... | 2023-06-07 |
| 64 | x11b | 6/20/2023 | 90 | 2 | 16 | Achillea millefolium (common yarrow), Astragal... | 2023-06-20 |
65 rows × 7 columns
# Split spp string into list of strings
bee = (
bee.
assign(
sp_list = lambda x: [i.split(", ") for i in x.sp_str]
)
)
bee.sort_values(['id', 'date'])
| id | date_str | survey_length | n_captured | n_flowering_plants | sp_str | date | sp_list | |
|---|---|---|---|---|---|---|---|---|
| 0 | x01a | 5/6/2023 | 45 | 0 | 17 | Alyssum desertorum (desert madwort), Arabis sp... | 2023-05-06 | [Alyssum desertorum (desert madwort), Arabis s... |
| 1 | x01a | 5/18/2023 | 90 | 2 | 22 | Alyssum desertorum (desert madwort), Astragalu... | 2023-05-18 | [Alyssum desertorum (desert madwort), Astragal... |
| 2 | x01a | 6/28/2023 | 90 | 1 | 26 | Achillea millefolium (common yarrow), Alyssum ... | 2023-06-28 | [Achillea millefolium (common yarrow), Alyssum... |
| 3 | x01b | 4/26/2023 | 45 | 0 | 6 | Alyssum desertorum (desert madwort), Lomatium ... | 2023-04-26 | [Alyssum desertorum (desert madwort), Lomatium... |
| 4 | x01b | 5/18/2023 | 90 | 0 | 28 | Allium textile (textile onion), Alyssum desert... | 2023-05-18 | [Allium textile (textile onion), Alyssum deser... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 60 | x10c | 6/20/2023 | 90 | 0 | 19 | Achillea millefolium (common yarrow), Alyssum ... | 2023-06-20 | [Achillea millefolium (common yarrow), Alyssum... |
| 61 | x11a | 4/23/2023 | 45 | 0 | 2 | Lomatium foeniculaceum (desert biscuitroot), P... | 2023-04-23 | [Lomatium foeniculaceum (desert biscuitroot), ... |
| 62 | x11b | 5/20/2023 | 90 | 0 | 11 | Allium textile (textile onion), Alyssum desert... | 2023-05-20 | [Allium textile (textile onion), Alyssum deser... |
| 63 | x11b | 6/7/2023 | 90 | 2 | 17 | Achillea millefolium (common yarrow), Alyssum ... | 2023-06-07 | [Achillea millefolium (common yarrow), Alyssum... |
| 64 | x11b | 6/20/2023 | 90 | 2 | 16 | Achillea millefolium (common yarrow), Astragal... | 2023-06-20 | [Achillea millefolium (common yarrow), Astraga... |
65 rows × 8 columns
# not quite right; the length of every sp_list shouldn't be 65...
[len(i) for i in bee.sp_list][:5]
# Ah. The sp_list is just the list of the flowering plants and will equal the n_flowering_plants col.
# it's not a list of all plants in the sampling area. Bummer.
[17, 22, 26, 6, 28]
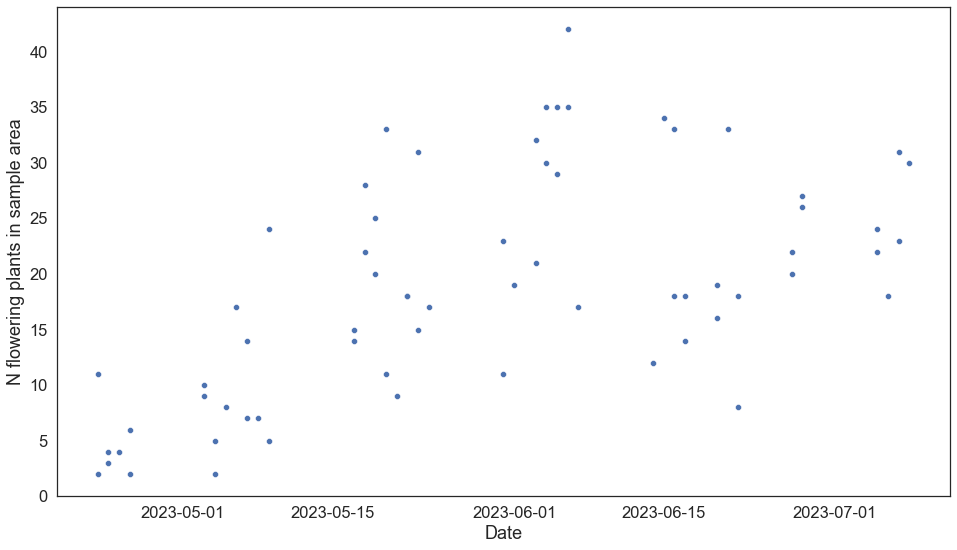
fig, ax = plt.subplots()
g = sns.scatterplot(data = bee, x = 'date', y = 'n_flowering_plants')
# g.axvline(35, color = 'black', ls = "--")
g.set(
ylabel = 'N flowering plants in sample area',
xlabel = 'Date'
)#xlim = (-25, 730))
[Text(0, 0.5, 'N flowering plants in sample area'), Text(0.5, 0, 'Date')]
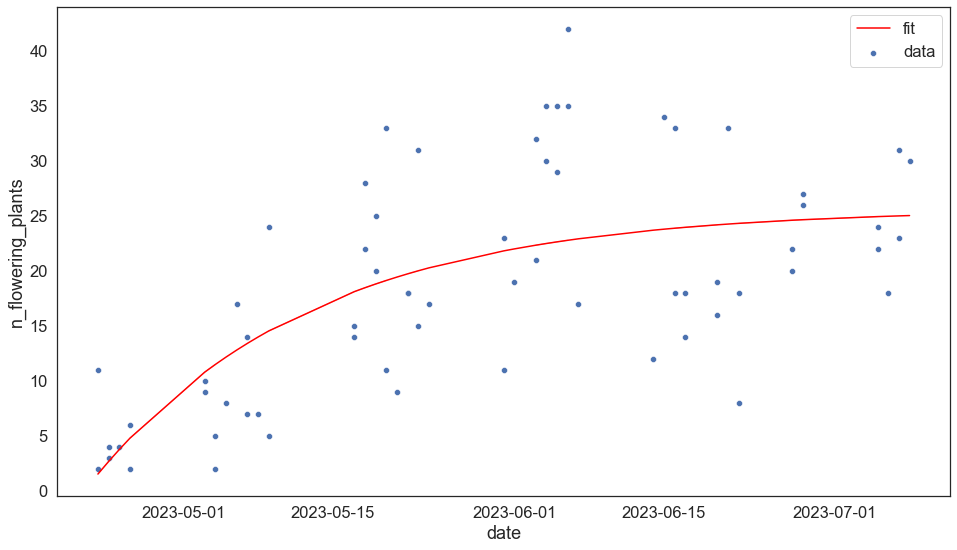
# Define the model
def von_b(x, Linf, K, t0):
return Linf * (1 - np.exp(-K*(x-t0)))
# optimized values, covariance of optimized values
params, cov = curve_fit(
f = von_b,
xdata = mdates.date2num(bee.date),
ydata = bee.n_flowering_plants,
p0 = [19, 0, 0]
)
print(params, "\n\n", cov)
[2.56409902e+01 4.85164279e-02 1.94687624e+04]
[[ 6.58188016e+00 -3.90031983e-02 -2.94344343e+00]
[-3.90031983e-02 3.19236318e-04 3.46377103e-02]
[-2.94344343e+00 3.46377103e-02 8.59442029e+00]]
xdata = mdates.date2num(bee.date)
ydata = bee.n_flowering_plants
fig, ax = plt.subplots()
# g = sns.scatterplot(xdata, von_b(xdata, *res), 'r-', label = 'fit')
sns.scatterplot(x = bee.date, y = ydata, label = 'data')
sns.lineplot(x = xdata, y = von_b(xdata, *params), c = 'red', label = 'fit')
# g # plt.show()
<AxesSubplot:xlabel='date', ylabel='n_flowering_plants'>
# Get CI, t-stat, and p-value for parameters from curve_fit()
# from: https://stats.stackexchange.com/questions/362520/how-to-know-if-a-parameter-is-statistically-significant-in-a-curve-fit-estimat
import scipy.odr
import scipy.stats
def f_wrapper_for_odr(params, x): # parameter order for odr
return von_b(x, *params)
model = scipy.odr.odrpack.Model(f_wrapper_for_odr)
data = scipy.odr.odrpack.Data(xdata, ydata)
myodr = scipy.odr.odrpack.ODR(data, model, beta0 = params, maxit = 0)
myodr.set_job(fit_type = 2)
parameterStatistics = myodr.run()
df_e = len(xdata)-len(params)
cov_beta = parameterStatistics.cov_beta
sd_beta = parameterStatistics.sd_beta * parameterStatistics.sd_beta
t_df = scipy.stats.t.ppf(0.975, df_e) #identify dof for 95% CI
ci = [] #Empty list for storage
for i in range(len(params)):
ci.append([params[i] - t_df * parameterStatistics.sd_beta[i], params[i] + t_df * parameterStatistics.sd_beta])
tstat_beta = params / parameterStatistics.sd_beta
pstat_beta = (1.0 - scipy.stats.t.cdf(np.abs(tstat_beta), df_e)) * 2.0
model.
<scipy.odr.odrpack.Model at 0x12fca0210>
ci
[[20.51049986684996, array([30.77148056, 25.67670011, 31.5033486 ])],
[0.012806529058685692, array([5.17900678, 0.08422633, 5.91087481])],
[19462.900041417906, array([19473.89289015, 19468.7981097 , 19474.62475818])]]
print(
params, "\n",
tstat_beta, "\n",
pstat_beta
)
for i in range(len(params)):
print('parameter:', params[i])
print(' conf interval:', ci[i][0], ci[i][1])
print(' t-statistic:', tstat_beta[i])
print(' p-value:', pstat_beta[i])
print()
[2.56409902e+01 4.85164279e-02 1.94687624e+04]
[9.99039188e+00 2.71585640e+00 6.63854015e+03]
[1.55431223e-14 8.55401612e-03 0.00000000e+00]
parameter: 25.640990214318908
conf interval: 20.51049986684996 [30.77148056 25.67670011 31.5033486 ]
t-statistic: 9.990391875787214
p-value: 1.554312234475219e-14
parameter: 0.04851642787630825
conf interval: 0.012806529058685692 [5.17900678 0.08422633 5.91087481]
t-statistic: 2.71585640409975
p-value: 0.00855401612286144
parameter: 19468.762399800315
conf interval: 19462.900041417906 [19473.89289015 19468.7981097 19474.62475818]
t-statistic: 6638.5401496559625
p-value: 0.0
import matplotlib.dates as mdates
mdates.date2num(bee.date)
array([19483., 19495., 19536., 19473., 19495., 19545., 19484., 19512.,
19545., 19484., 19512., 19536., 19471., 19499., 19535., 19481.,
19498., 19525., 19473., 19508., 19530., 19496., 19511., 19546.,
19486., 19500., 19535., 19494., 19509., 19530., 19485., 19500.,
19522., 19481., 19499., 19523., 19470., 19501., 19525., 19486.,
19513., 19544., 19496., 19513., 19529., 19472., 19497., 19524.,
19480., 19514., 19543., 19480., 19514., 19543., 19482., 19508.,
19524., 19471., 19494., 19511., 19528., 19470., 19497., 19515.,
19528.])
# import numpy as np
# from scipy.optimize import curve_fit
# import matplotlib.pyplot as plt
# def f(x, start, end):
# res = np.empty_like(x)
# res[x < start] =-1
# res[x > end] = 1
# linear = np.all([[start <= x], [x <= end]], axis=0)[0]
# res[linear] = np.linspace(-1., 1., num=np.sum(linear))
# return res
# if __name__ == '__main__':
# xdata = np.linspace(0., 1000., 1000)
# ydata = -np.ones(1000)
# ydata[500:1000] = 1.
# ydata = ydata + np.random.normal(0., 0.25, len(ydata))
# popt, pcov = curve_fit(f, xdata, ydata, p0=[495., 505.])
# print(popt, pcov)
# plt.figure()
# plt.plot(xdata, f(xdata, *popt), 'r-', label='fit')
# plt.plot(xdata, ydata, 'b-', label='data')
# plt.show()
np.empty_like(bee.date)
array(['2118-01-08T11:31:00.216823808', '2118-01-08T12:25:58.751707136',
'2118-01-08T15:33:48.745891840', '2118-01-08T10:45:11.437754368',
'2118-01-08T12:25:58.751707136', '2118-01-08T16:15:02.647054336',
'2118-01-08T11:35:35.094730752', '2118-01-08T13:43:51.676125184',
'2118-01-08T16:15:02.647054336', '2118-01-08T11:35:35.094730752',
'2118-01-08T13:43:51.676125184', '2118-01-08T15:33:48.745891840',
'2118-01-08T10:36:01.681940480', '2118-01-08T12:44:18.263334912',
'2118-01-08T15:29:13.867984896', '2118-01-08T11:21:50.461009920',
'2118-01-08T12:39:43.385427968', '2118-01-08T14:43:25.088915456',
'2118-01-08T10:45:11.437754368', '2118-01-08T13:25:32.164497408',
'2118-01-08T15:06:19.478450176', '2118-01-08T12:30:33.629614080',
'2118-01-08T13:39:16.798218240', '2118-01-08T16:19:37.524961280',
'2118-01-08T11:44:44.850544640', '2118-01-08T12:48:53.141241856',
'2118-01-08T15:29:13.867984896', '2118-01-08T12:21:23.873800192',
'2118-01-08T13:30:07.042404352', '2118-01-08T15:06:19.478450176',
'2118-01-08T11:40:09.972637696', '2118-01-08T12:48:53.141241856',
'2118-01-08T14:29:40.455194624', '2118-01-08T11:21:50.461009920',
'2118-01-08T12:44:18.263334912', '2118-01-08T14:34:15.333101568',
'2118-01-08T10:31:26.804033536', '2118-01-08T12:53:28.019148800',
'2118-01-08T14:43:25.088915456', '2118-01-08T11:44:44.850544640',
'2118-01-08T13:48:26.554032128', '2118-01-08T16:10:27.769147392',
'2118-01-08T12:30:33.629614080', '2118-01-08T13:48:26.554032128',
'2118-01-08T15:01:44.600543232', '2118-01-08T10:40:36.559847424',
'2118-01-08T12:35:08.507521024', '2118-01-08T14:38:50.211008512',
'2118-01-08T11:17:15.583102976', '2118-01-08T13:53:01.431939072',
'2118-01-08T16:05:52.891240448', '2118-01-08T11:17:15.583102976',
'2118-01-08T13:53:01.431939072', '2118-01-08T16:05:52.891240448',
'2118-01-08T11:26:25.338916864', '2118-01-08T13:25:32.164497408',
'2118-01-08T14:38:50.211008512', '2118-01-08T10:36:01.681940480',
'2118-01-08T12:21:23.873800192', '2118-01-08T13:39:16.798218240',
'2118-01-08T14:57:09.722636288', '2118-01-08T10:31:26.804033536',
'2118-01-08T12:35:08.507521024', '2118-01-08T13:57:36.309846016',
'2118-01-08T14:57:09.722636288'], dtype='datetime64[ns]')
And now for something completely different
Lets look at the other data file in the .zip
bee = pd.read_csv('../../Google Drive/job_tech_learning/data/Bumblebeesandfl/MT_2023_Plant_Species.csv')
bee.head()
| Genus | Species | Common Name | |
|---|---|---|---|
| 0 | Acer | negundo | boxelder |
| 1 | Achillea | millefolium | common yarrow |
| 2 | Agoseris | glauca | pale agoseris |
| 3 | Allium | cernuum | nodding onion |
| 4 | Allium | textile | textile onion |